The Improvement of African Orphan Crops through TILLING
Korinna Esfeld and Zerihun Tadele
Institute of Plant Sciences, University of Bern, Altenbergrain 21, 3013 Bern, Switzerland
(Email: korinna.esfeld@ips.unibe.ch, zerihun.tadele@ips.unibe.ch)
Abstract
People in African countries mostly depend on special staple crops – orphan crops – that are particularly important for their food security, nutrition and income. These crops are better adapted to local soil and climatic conditions as well as to the agro-ecology and socio-economic conditions in developing countries. However major challenges in orphan crops are low productivity, low nutrition and the production of toxic substances. Conventional breeding methods often do not overcome these problems and transgenic methods can not be applied due to several reasons. Alternative molecular methods like TILLING (Targeting Induced Local Lesion IN Genomes) are needed from which orphan crops may benefit. TILLING is a general, easy, non-transgenic and low-cost reverse genetic method to identify single base pair changes in genes of choice. The technique was first developed for the model plant Arabidopsis but successfully adopted to other species including crop plants. The procedure of TILLING comprises: classical mutagenesis, development of a non-chimeric population, preparation of a germplasm stock, DNA extraction and sample pooling, population screening for induced mutations as well as validation and evaluation of candidates. Here, we present a general overview of the TILLING method with special focus on crops and give an example how the technique can also be applied easily to orphan crops. We discuss our experience on tef (Eragrostis tef), one of the understudied crops of Africa.
Key words: Orphan crops, TILLING, EcoTILLING, mutagenesis, Eragrostis tef
Need for orphan crops improvement
Feeding the ever-increasing population of Africa will be a challenge in the future. Most of the population of the continent depend as food source or income generation on so called ‘orphan crops’ which are mainly unknown outside their countries or have at least no economic importance. However, these crops are particularly important for food security, nutrition and income to resource-poor farmers and consumers in developing countries. In addition, orphan crops are much better adapted to the often difficult local soil and climatic conditions. Although large number of orphan crops are known to exist on the continent, the major ones are cereals [e.g., finger millet (Eleusine coracana), tef (Eragrostis tef) and fonio (Digitaria spp)], legumes [cowpea (Vigna unguiculata), bambara groundnut (Vigna subterranea) and grass pea (Lathyrus sativus)] and root crops [cassava (Manihot esculenta), yam (Dioscorea spp.) and enset (Ensete ventricosum)]. Despite their importance in adapting to the adverse agro-ecological conditions, orphan crops have also several limitations. Some of the prominent bottlenecks related to these crops are low productivity (e.g. in tef), poor in essential nutrients (cassava and enset) or production of toxic substances (cassava and grass pea; for review [1]). Another challenge is that breeders of orphan crops are mostly depend on the conventional breeding techniques particularly on selection which fail to improve some valuable traits in these crops. Modern biotechnological techniques including the transgenic approach are not yet employed for orphan crops due to negative perception in most African countries and the lack of biosafety regulations.
TILLING has high potential to improve orphan crops
Various types of crop improvement techniques are known to exist. Broadly they are grouped into two although sometimes there is no clear distinction between these two: i) conventional techniques that mainly include various types of selection methods and introgressions or hybridizations, ii) biotechnological or modern techniques that include transgenic and non-transgenic techniques. Among non-transgenic methods TILLING (Targeting Induced Local Lesion IN Genomes) becomes recently popular and is extensively implemented for major crops and to a certain level to orphan crops. TILLING is a general, easy, non-transgenic and low-cost reverse genetic method which uses traditional mutagenesis followed by high-throughput mutation detection. It identifies single base pair changes in targeted genes and can be applied to every organism independent of the genome size, reproductive system, generation time and polyploidy level [2, 3, 4, 5, 6, 7]. In contrast to forward genetic screenings where the mutants are first selected based on the phenotype reverse genetic approaches refer to the targeted discovery of mutations in genes known by their sequence [5].
TILLING was first developed and established in the model plant Arabidopsis thaliana [2] but was later successfully adopted to numerous animal and plant species. The technique is so far implemented in crops such as pea, soybean, maize, barley, rice, wheat, sorghum and in the orphan crop tef [8, 9, 10]. Thus, since its first description in the year 2000 [2] TILLING gained a lot of popularity [9]. TILLING can also be useful in inducing a wider genetic diversity in the genomes of domesticated species. Classical breeding approaches such as domestication and selection are facing the problem of limited genetic diversity in adapted lines since much of the genetic variation available in wild crop progenitors has been lost. TILLING can introduce genetic variation directly to elite germplasms without the need to acquire variation from exotic cultivars avoiding the introduction of agriculturally undesirable traits. In this case, several backcrosses to the parent cultivar remove unlinked mutations and result in a novel allele in the parental background [11, 12].
A general overview of the TILLING method is given in Fig. 1. Till et al. [13] present a simple and efficient protocol of the technique. In general the technique of TILLING comprises the following main steps: i) mutagenesis, ii) development of a non-chimeric population, iii) preparation of a germplasm stock, iv) DNA extraction and sample pooling, v) screening of the population for induced mutations as well as validation and vi) evaluation of the candidates [7, 9].
In addition, TILLING can also be applied to detect naturally occurring single base changes known as SNP (Single Nucleotide Polymorphism) that correspond to the randomly induced mutations. This adapted method is known as EcoTILLING [14].
Mutagenesis is the crucial step in TILLING process
The starting point of TILLING is the mutagenesis of seeds or pollen to induce single nucleotide changes [11]. Conventional chemical mutagenesis has a long history in crop breeding and the broad experience simplifies its application [4, 7, 9, 12, 15]. The technique of TILLING can be applied to nearly all species even to orphan crops that lack well developed genetic tools [16]. In addition since no exogenous DNA is introduced into the plant, the technique is considered as non-transgenic and the products are exempted from regulatory restrictions that are imposed on the transgenic products [11, 15, 16].
A number of mutagens are used to create mutations in different organisms. Broadly the mutagens are grouped into two: i) chemical mutagens including ethyl methanesulfonate (EMS), sodium azide, N-methyl-N-nitrosourea (MNU), methyl methanesulfonate (MMS), hydrogen fluoride (HF) and hydroxylamine, and ii) physical mutagens such as gamma- and x-ray. Mutations are randomly induced and target virtually every gene depending on the mutation density [8, 17]. The advantage of chemical mutagenesis is that it creates an allelic series of mutations. Three types of mutations are recovered from chemical mutagenesis: i) truncation or nonsense mutation: where a single base pair change coverts an amino acid codon into a stop codon, ii) missense mutation: in which a single base pair change alters the amino acid encoded by a particular codon and can be distinct into conservative and non-conservative changes and iii) silent mutation: where a single base pair does not alter the amino acid encoded by a particular codon [2]. EMS is the most commonly used mutagen especially for TILLING experiments because it induces point mutations [15]. It specifically creates a G:C to A:T transitions since it alkylates G residues which then pair with T instead of C [7, 8]. On average, in the genome mutagenized with EMS, the following mutation rates are expected: 3% truncations, 50% missense and 48% silent mutations [16].
The allelic series of induced mutations can potentially confer to various phenotypes that range from subtle to strong. Mutations in the coding region of the gene might alter plant metabolism and maybe the effective level of a gene product that might be useful for breeding. In addition, splice site mutations that inhibit proper intron splicing, partial loss-of-function as well as novel-function alleles may occur [5, 9, 11].
The most important point to be considered while making mutagenesis is to balance between the mutation density and a feasible germination rate that is also linked to low sterility of the plants after mutagenesis [8, 18, 19]. Therefore, pilot studies need to be made before embarking large-scale mutagenesis in order to find the right mutagen, optimum concentration and proper handling of the chemical [9, 19]. In general, optimizing the concentration of the mutagen is difficult for diploid species since these plants have lower tolerance towards mutagens; hence increases the amount of mutagenized population to be screened [8]. On the other hand, polyploids show a higher tolerance due to complementation of essential genes by homeologous copies; therefore, expected mutations could be revealed from smaller sized populations [8]. However, in polyploid species genetic buffering makes it less likely that recessive mutations show a phenotype. Therefore, it may be necessary to identify mutations in each homeologous copy of the targeted gene and bring these together by crossing [8].
The mutation rate is estimated as the total number of mutations scored divided by the total number of base pairs screened, i.e. amplicon size x screened individuals [16]. According to Weil [10] one mutation per 500 kb or less is regarded as optimal. The highest mutation density was obtained from two polyploid wheats [18] i.e., one mutation per 25 kb in hexaploid wheat and one mutation per 40 kb in tetraploid wheat as compared to one mutation per 500 kb in maize and rice [7].
Detailed procedures of the TILLING method
Although some minor procedural differences are reported from various labs depending on the nature of plants and availability of resources, most TILLING experiments apply the following basic four steps.
1. Mutagenesis
For the majority of plant species, with the exception of maize, seeds are used as source of mutagenesis [7, 15]. Due to the multicellular stage of embryos in seeds, the first generation of mutagenized plants (defined as M1 population) is typically chimeric; i.e., different cells make different genotypes [9]. Hence, M1 plants are selfed and a single seed from each M1 plant is used to establish the M2 population. In comparison to the M1 plants, plants growing from M2 seeds are uniform, do not segregate in their cells and the induced changes are stable and heritable [15, 16]. Tissue samples are collected for DNA extraction from individual M2 plants while the seeds are harvested and long time stored from the M3 population [4].
2. DNA sampling
Genomic DNA is extracted from the tissues collected from individual M2 lines and subsequently normalized to achieve identical DNA concentrations for all samples. This will be followed by pooling of DNA samples from 2x up to 8x in a one- or two dimensional range. In addition to saving the cost and time of the screening, pooling also facilitates the detection of potential mutations [9, 15]. Two dimensional pooling has additional advantage in avoiding false positives since candidate mutants are visualized at two independent sites [9].
3. PCR amplification
PCR amplification is performed using a set of Infra-red dye (IRD) labelled specific primers for the gene of choice. The gene of choice refers to the gene that regulates the traits of interest. Even though prior information on the genome sequence is not required for TILLING, the presence of full-length genomic sequence for the gene of interest improves the chance of success and accelerates the development of suitable targets especially in designing effective primers [4, see also 8]. Specificity of the primers is important especially when various members of gene families are needed to be amplified and particularly in polyploid species [9, 11, 18]. Specificity of primers can be improved in polyploid species focusing on more divergent regions particularly in the intron region [12, 18]. Another approach to tackle the problem of gene copies in polyploid species is to pre-treat the genomic DNA prior to PCR amplification with a restriction enzyme that removes only one copy [16].
4. Mutation detection
PCR products amplified using fluorescent labelled primers are separated on a denaturating polyacrylamide gel for detection of mutations. In order to increase the efficiency of PCR amplification, unlabelled primers are added in the same PCR reaction with the labelled ones. The length of the amplified products could range between 0.3 to 1.6 kb [11]. However, earlier studies reported that it is difficult to detect mutations at the ends of fragments and normally 200 bp have to be excluded from the analysis; 100 bp from each side [19].
The PCR amplification is followed by the heteroduplex formation step where the amplified products are first denatured and then slowly cooled. In this step, heteroduplex molecules are formed due to the mismatches that occur when wild type and mutant DNA anneal [3, 6]. While at the beginning detection of single base pair differences were done using denaturating HPLC (DHPLC; [2]) nowadays single-strand cleavage is used to detect mutations [4, 6, 15]. The mismatches or heteroduplexes are recognized and cleaved by single-strand specific nucleases that are members of the S1 nuclease family such as CEL I and mung bean nuclease [7, 20]. CEL I is isolated from celery leaves and it is the most preferred enzyme for mutation detection in TILLING projects [13, 20]. The CEL I enzyme cleaves to the 3’ side of mismatches and loop outs in heteroduplexes while leaving duplexes intact [4]. The CEL I digested products are purified (e.g. using Sephadex ® purification; [13]). Finally products are resolved on denaturating polyacrylamide gels. LI-COR DNA analyzer is the standard system for most TILLING projects.
Since forward and reverse primers are labelled with different infrared dyes, cleaved products are visible in both channels of a LI-COR machine (Fig. 2). Confirmation for the right mutation detection is made when the two cleaved products observed in different channels sum up to the original PCR fragment size. Another important advantage of the mismatch cleavage is that it pinpoints to the location of the polymorphism making confirmation by sequencing quite efficient [6, 15].
Variations in mutation detection can also be done, for example, using capillary electrophoresis [6, 8] and recently agarose gels [8, 21].
Web-based tools applied in TILLING
Several web-based and freely available programs facilitate various operations in TILLING procedure. Some of the widely used tools are indicated below.
.CODDLE (Codons Optimized to Detect Deleterious Lesions; http://www.proweb.org/coddle/; accessed April 2010) allows to target a functional domain or the domain which is likely to be the most sensible to amino-acid substitutions (Fig. 3). The use of this program increases the probability of: i) detecting deleterious mutation in the gene of interest, and ii) obtaining regions with high frequency of stop codons and those which are evolutionary conserved hence useful for providing an allelic series [17].
.GelBuddy (http://www.proweb.org/gelbuddy/installissues.html; accessed April 2010) is used to automate band calling in the electrophoretic gels [13].
.PARSESNP (Project Aligned Related Sequences and Evaluate SNPs; http://www.proweb.org/parsesnp/; accessed April 2010) is useful in revealing the changes in the nucleotide and amino-acid sequences as well as documenting any restriction endonuclease site that have been altered [6].
.SIFT (http://sift.jcvi.org/; accessed April 2010) is useful in predicting whether the change in the amino-acid has deleterious effect on the protein [4, 22].
Application of TILLING to the orphan crop improvement
To date, the technique of TILLING and/or EcoTILLING is applied to few orphan crops including cassava, banana and tef [9]. The cassava and banana projects are based at the Joint FAO/IAEA Program in Vienna while the Tef TILLING and EcoTILLING Projects are hosted by the Institute of Plant Sciences, University of Bern, Switzerland. Tef is a major cereal crop from Ethiopia that is closely related to finger millet. Similar to other orphan crops, tef adapts to diverse climatic and soil conditions and also tolerates many pests and diseases. In addition to its nutritional advantages, seeds of tef are also free of gluten for which a high number of people are allergic. However, the productivity of tef is limited mainly due to the prevalence of lodging. The tef plant has a tall and tender stem which is susceptible to damage by wind and rain. Therefore, developing lodging resistant semi-dwarf tef cultivars is the main goal of the Tef TILLING and EcoTILLING Projects. About 7000 EMS mutagenized M2 population plants and 500 accessions are generated for the TILLING and EcoTILLING, respectively. Genes known to control plant height in major crop species including the so called ‘Green Revolution genes’ of rice and wheat are used as a target. The current identification of a number of genes affecting plant height from major cereals crops including wheat, rice and maize (for review [23]) facilitate TILLING of plant height genes in tef. In addition, the information from the Tef Genome Sequencing Initiative (see this issue [24]) improves the application of TILLING in tef. Full-length genomic clones for two genes were isolated from tef. The two genes are homologs of the DWARF 4 gene [25] and the HIGH TILLERING and DWARF 1 gene from rice, respectively (HTD1, [26]).
One of the major obstacle in our Tef TILLING Project is related to the polyploidy nature of tef, as the species is allotetraploid, that is, each gene exists in two copies. In order to detect intended mutations, only a single copy should be amplified. This particular polyploidy problem is overcome by designing at least one of the two primers from the unique intron region in order to amplify a single copy at a time.
The detailed procedure we adopted for our Tef TILLING project is briefly indicated below:
.DNA extraction using the Machery-Nagel NucleoSpin® 96 Plant extraction kit
.DNA normalization to 5 ng and 4x pooling in a two dimensional range
.Amplification of target genes (single step PCR using GoTaq polymerase; Promega); PCR products vary between 770 bp-1140 bp
.Heteroduplex formation and digestion of mismatches with the CEL I enzyme
.Separation on denaturating polyacrylamide gels (LI-COR DNA analyzer)
.Detection and sequencing of the candidates as well as phenotypic confirmation
So far, by screening 3264 mutants using the DWARF 4 gene, four candidates were recovered. Similarly, by screening 4224 plants for the HTD 1 gene, 12 candidates were obtained. These initial results show that the frequency of mutation in tef for the two genes is one mutation per 465 kb for DWARF 4 and one mutation per 348 kb for HTD1. The next steps are to further investigate the phenotype of the candidate lines and to screen more mutants using other plant height-related genes.
Summary and future perspectives
TILLING is a general, easy and non-transgenic reverse genetic method that was first described for Arabidopsis thaliana. Quite rapidly the method was adopted to several plant and animal species including widely cultivated crops such as rice, wheat and maize. In addition, there is a growing interest to apply TILLING to orphan crops that lack well developed genetic tools. The technique is proved to be efficient in obtaining desirable mutant lines of agronomic importance. However, before new cultivars will be released to the farming community a number of backcrosses (at least four) should be done to remove undesirable traits [9, 11].
Acknowledgements
We would like to express our gratitude to the Syngenta Foundation for Sustainable Agriculture and to the University of Bern for financially and technically supporting our project on Tef TILLING and EcoTILLING.
References
1. Tadele, Z. (ed; 2009) New approaches to plant breeding of orphan crops in Africa: Proceedings of an International Conference, 19-21 September 2007, Bern, Switzerland. Stämpfli AG, Bern.
2. McCallum, C. M., Comai, L., Greene, E. A. and Henikoff, S. (2000) Targeted screening for induced mutations. Nature Biotechnology 18, 455-457.
3. Till, B. J., Reynolds, S. H., Greene, E. A., Codomo, C. A., Enns, L. C., Johnson, J. E., Burtner, C., Odden, A. R., Young, K., Taylor, N. E., Henikoff, J. G., Comai, L. and Henikoff, S. (2003) Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Research 12, 524-530.
4. Henikoff, S., Till, B. J. and Comai, L. (2004) TILLING. Traditional mutagenesis meets functional genomics. Plant Physiology 135, 1-7.
5. Stemple, D. L. (2004) TILLING – a high-throughput harvest for functional genomics. Genetics 5, 1-7.
6. Gilchrist, E. J. and Haughn, G. W. (2005) TILLING without a plough: a new method with applications for reverse genetics. Current Opinion in Plant Biology 8, 211-215.
7. Comai, L. and Henikoff, S. (2006) TILLING: practical single-nucleotide mutation discovery. The Plant Journal 45, 684-694.
8. Parry, M. A., Madgwick, P. J., Bayon, C., Tearall, K., Hernandez-Lopez, A., Baudo, M., Rakszegi, M., Hamada, W., Al-Yassin, A., Ouabbou, H., Labhilili, M. and Phillips, A. L. (2009) Mutation discovery for crop improvement. Journal of Experimental Botany 66, 2817-2825.
9. Tadele, Z., Mba, C. and Till, B. J. (2009) TILLING for mutations in model plants and crops. In S. Mohan Jain and D. S. Brar (eds.) Molecular Techniques in Crop Improvement. Springer Netherlands.
10. Weil, C. F. (2009) TILLING in grass species. Plant Physiology 149, 158-164.
11. Slade, A. J. and Knauf, V. C. (2005) TILLING moves beyond functional genomics into crop improvement. Transgenic Research 14, 109-115.
12. Sestili, F., Botticella, E., Bedo, Z., Phillips, A. and Lafiandra, D. (2010) Production of novel allelic variation for genes involved in starch biosynthesis through mutagenesis. Molecular Breeding 25, 145-154.
13. Till, B. J., Zerr, T., Comai, L. and Henikoff, S. (2006) A protocol for TILLING and Ecotilling in plants and animals. Nature Protocols 1, 2465-2477.
14. Comai, L., Young, K., Till, B. J., Reynolds, S. H., Greene, E.A., Codomo, C. A., Enns, L. C., Johnson, J. E. , Burtner, C., Odden, A. R. and Henikoff, S. (2004) Efficient discovery of DNA polymorphisms in natural populations by ecotilling. Plant Journal 37, 778-786.
15. Till, B. J., Comai, L. and Henikoff, S. (2007) TILLING and EcoTILLING for crop improvement. In R. K. Varshney and R. Tuberosa Genomics-Assisted Crop Improvement,
Vol. 1: Genomics Approaches and Platforms. Springer, Netherlands.
16. Cooper, J. L., Till, B. J., Laport, R. G., Darlow, M. C., Kleffner, J. M., Jamai, A., El-Mellouki, T., Liu, S., Ritchie, R., Nielsen, N., Bilyeu, K. D., Meksem, K., Comai, L. and Henikoff, S. (2008) TILLING to detect induced mutations in soybean. BMC Plant Biology 8, doi:10.1186/1471-2229-8-9.
17. Colbert, T., Till, B. J., Tompa, R., Reynolds, S., Steine, M. N., Yeung, A. T., McCallum, C. M., Comai, L. and Henikoff, S. (2001) High-throughput screening for induced point mutations. Plant Physiology 126, 480-484.
18. Slade, A. J., Fuerstenberg, S. I., Loeffler, D., Steine, M. N. and Facciotti, D. (2005) A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nature Biotechnology 23, 75-81.
19. Xin, Z., Wang, M. L., Barkley, N. A., Burow, G., Franks, C., Pederson, G. and Burke, J. (2008) Applying genotyping (TILLING) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biology 8, 103-116.
20. Till, B. J., Burtner, C., Comai, L. and Henikoff, S. (2004) Mismatch cleavage by single-strand specific nucleases. Nucleic Acids Research 32, 2632-2641.
21. Uauy, C., Paraiso, F., Colasuonno, P., Tran, R., Tsai, H., Berardi, S., Comai, L. and Dubcosvky, J. (2009) A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biology, 9:115, doi:10.1186/1471-2229-9-115.
22. Ng, P. C. and Henikoff, S. (2003) SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Research 31, 3812-3814.
23. Wang, Y. and Li, J. (2006) Genes controlling plant architecture. Current Opinion in Biotechnology 17, 123-129.
24. Plaza, S., Bossolini, E., and Tadele, Z. (2010) Significance of Genome Sequencing for African Orphan Crops: the case of Tef (this issue).
25. Sakamoto, T., Morinaka, Y., Ohnishi, T., Sunohara, H., Fujioka, S., Ueguchi-Tanaka, M., Mizutani, M., Sakata, K., Takatsuto, S., Yoshida, S., Tanaka, H., Kitano, H. and Matsuoka, M. (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nature Biotechnology 24, 105-109.
26. Zou, J., Chen, Z., Zhang, S., Zhang, W., Jiang, G., Zhao, X., Zhai, W., Pan, X. and Zhu, L. ( 2005) Characterizations and fine mapping of a mutant gene for high tillering and dwarf in rice (Oryza sativa L.). Plant 222:604-612.
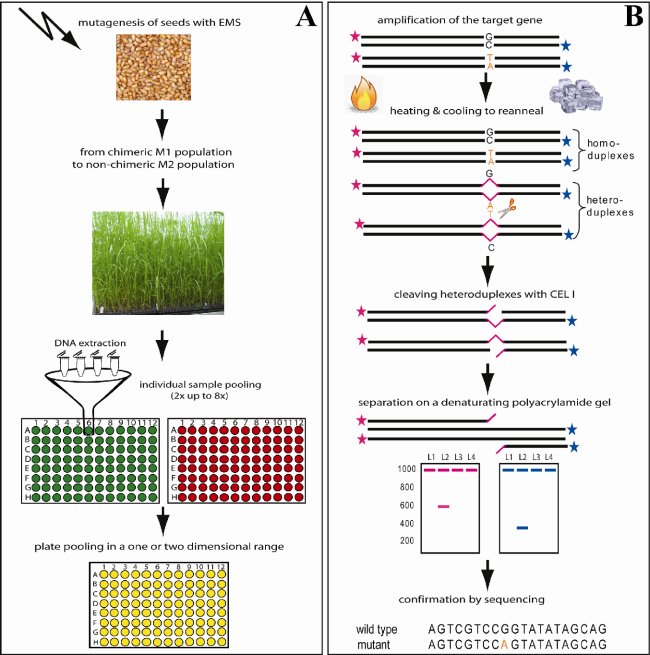
Fig. 1 shows the diagrammed TILLING procedure. Panel A) starts with the beginning of the TILLING process, the mutagenesis of the seeds with EMS. Subsequently the seeds are grown to chimeric M1 plants. These plants are selfed and a single seed of each plant is used to establish the screening population (M2 population). M3 seeds are used for further investigation. Tissue samples from M2 plants are used for the DNA extraction. DNA samples are normalized to assure equal concentrations and subsequently pooled 2x up to 8x in a one- or two dimensional range. The example above shows 4x pooling in a two dimensional range. After preparation of the DNA samples the amplification of specific target genes follows with specific labelled and unlabelled primer combinations (Panel B). The amplified DNA strands are then denatured by heating and slowly cooled down to allow heteroduplex formation. Mismatches that occur in heteroduplex molecules are cleaved with the enzyme CEL I. Finally the purified PCR products are separated on denaturating polyacrylamide gels for mutation detection. Cleaved PCR products are visible each time in one of the two channels of a LI-COR DNA analyzer and sum up to the full-length fragment size. The last step shows the confirmation of candidates by sequencing and phenotypic examination (not shown).
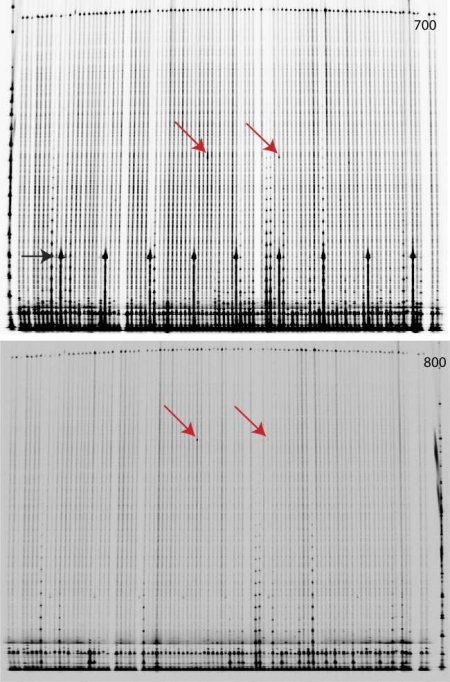
Fig. 2. Discovery of induced mutations in Eragrostis tef by TILLING. Individual DNA samples of mutated E. tef plants (M2 population) were pooled 4x in a two dimensional range. The target region was amplified using a specific pair of infra-red labelled (IRDye 700 and 800) and unlabelled primers. After heteroduplex formation, samples were digested with the CEL I enzyme purified from celery juice. Mismatches were cleaved and samples were separated on a denaturating polyacrylamide gel after Sephadex purification. Gel electrophoresis was performed using a LI-COR DNA analyzer. For each gel run, two images were produced, one for DNA labelled with IRDye 700 (top) and one for IRDye 800 (bottom). The red arrows indicate cleaved PCR products and the molecular weight of the respective cleaved fragments detected in the 700 and 800 channel of the LI-COR analyzer sum up to the molecular weight of the full-length PCR product size. Due to the two dimensional pooling the two detected mutations refer to a single plant. The black arrow indicates a labelled 200 bp fragment that was applied to the polyacrylamide gel every 10th lane to facilitate the lane scoring. The analysis was done with help of GelBuddy software.
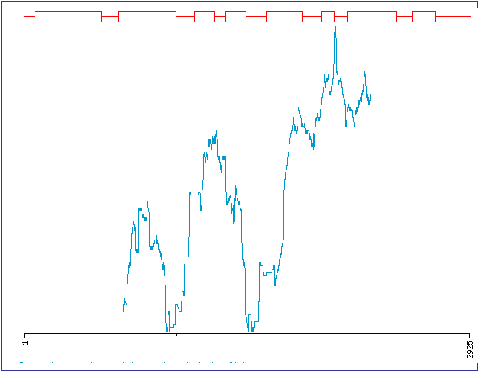
Fig. 3. CODDLE analysis of the tef HTD1 gene (EtHTD1). In the above example the CODDLE program uses the genomic and cDNA sequence of the EtHTD1 gene to determine exons which bring nonsense and missense mutations when mutagenized by EMS. Since COODLE selected exons 5, 6 and 7 as high potential regions for the two types of mutations, our screening using TILLING focused on this part of the HTD1 gene. CODDLE is freely available at http://www.proweb.org/coddle/